beginner
Reputation: 411
How to remove character with specific pattern form data frame in R
Im trying to remove the all the characters starting with the pattern "Gm" from last column of my data.frame
My data.frame looks like this
level logp chr start end CNA Genes
3 1.4 3 100 110 gain Gm5852,Gm5773,Tdpoz4,Tdpoz3,Gm911
4 18.10 3 962 966 gain Fcgr1,Terc,Gm5703
The result should look something like this
level logp chr start end CNA Genes
3 1.4 3 100 110 gain Tdpoz4,Tdpoz3
4 18.10 3 962 966 gain Fcgr1,Terc
Upvotes: 3
Views: 896
Answers (2)
G. Grothendieck
Reputation: 270248
This uses a single gsub to remove the unwanted portions:
Genes <- c("Gm5852,Gm5773,Tdpoz4,Tdpoz3,Gm911", "Fcgr1,Terc,Gm5703") # test data
gsub(",?Gm[^,]*,?", "", Genes)
giving:
[1] "Tdpoz4,Tdpoz3" "Fcgr1,Terc"
Here is a visualization of the regular expression:
,?Gm[^,]*,?
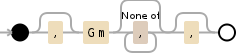
Upvotes: 5

Marat Talipov
Reputation: 13314
Given a data frame d:
d$Genes_new <- sapply(strsplit(as.character(d$Genes),split=','),function(s) paste(s[!grepl('^Gm',s)],collapse=','))
# level logp chr start end CNA Genes Genes_new
#1 3 1.4 3 100 110 gain Gm5852,Gm5773,Tdpoz4,Tdpoz3,Gm911 Tdpoz4,Tdpoz3
#2 4 18.1 3 962 966 gain Fcgr1,Terc,Gm5703 Fcgr1,Terc
Here, strsplit(as.character(d$Genes),split=',') creates a list of comma-separated gene names for each row, and sapply applies to each element of this list a function that excludes all gene names starting from Gm (s[!grepl('^Gm',s)]) and concatenates the remaining genes (paste(.,collapse=',').
Upvotes: 2
Related Questions
- How to remove specific characters from string in a column in R?
- remove a character from the entire data frame
- How to remove certain characters from a dataframe in R?
- How do I remove specific characters from a data frame
- How to remove matching characters in R?
- Remove some value (that contain a specific character) in a column
- How to remove a character in specific columns in a data.frame with R?
- Removing character strings from dataframe in R
- Removing character from dataframe
- remove some characters in a data.frame in R