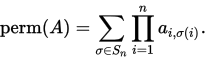Reputation: 21343
Can this cython code be optimized?
I am using cython for the first time to get some speed for a function. The function takes a square matrix A floating point numbers and outputs a single floating point number. The function it is computing is the permanent of a matrix
When A is 30 by 30 my code takes about 60 seconds currently on my PC.
In the code below I have implemented the Balasubramanian-Bax/Franklin-Glynn formula for the permanent from the wiki page. I have called the matrix M.
One sophisticated part of the code is the array f which is used to hold the index of the next position to flip in the array d. The array d holds values that are +-1. The manipulation of f and j in the loop is just a clever way to update a Gray code quickly.
from __future__ import division
import numpy as np
cimport numpy as np
cimport cython
DTYPE_int = np.int
ctypedef np.int_t DTYPE_int_t
DTYPE_float = np.float64
ctypedef np.float64_t DTYPE_float_t
@cython.boundscheck(False) # turn off bounds-checking for entire function
@cython.wraparound(False) # turn off negative index wrapping for entire function
def permfunc(np.ndarray [DTYPE_float_t, ndim =2, mode='c'] M):
cdef int n = M.shape[0]
cdef np.ndarray[DTYPE_float_t, ndim =1, mode='c' ] d = np.ones(n, dtype=DTYPE_float)
cdef int j = 0
cdef int s = 1
cdef np.ndarray [DTYPE_int_t, ndim =1, mode='c'] f = np.arange(n, dtype=DTYPE_int)
cdef np.ndarray [DTYPE_float_t, ndim =1, mode='c'] v = M.sum(axis=0)
cdef DTYPE_float_t p = 1
cdef int i
cdef DTYPE_float_t prod
for i in range(n):
p *= v[i]
while (j < n-1):
for i in range(n):
v[i] -= 2*d[j]*M[j, i]
d[j] = -d[j]
s = -s
prod = 1
for i in range(n):
prod *= v[i]
p += s*prod
f[0] = 0
f[j] = f[j+1]
f[j+1] = j+1
j = f[0]
return p/2**(n-1)
I have used all the simple optimizations I found in the cython tutorial. Some aspects I have to admit I don't fully understand. For example, if I make the array d ints, as the values are only ever +-1, the code runs about 10% more slowly so I have left it as float64s.
Is there anything else I can do to speed up the code?
This is the result of cython -a . As you can see everything in the loop is compiled to C so the basic optimizations have worked.
Here is the same function in numpy which is over 100 times slower than my current cython version.
def npperm(M):
n = M.shape[0]
d = np.ones(n)
j = 0
s = 1
f = np.arange(n)
v = M.sum(axis=0)
p = np.prod(v)
while (j < n-1):
v -= 2*d[j]*M[j]
d[j] = -d[j]
s = -s
prod = np.prod(v)
p += s*prod
f[0] = 0
f[j] = f[j+1]
f[j+1] = j+1
j = f[0]
return p/2**(n-1)
Timings updated
Here are timings (using ipython) of my cython version, the numpy version and romeric's improvement to the cython code. I have set the seed for reproducibility.
from scipy.stats import ortho_group
import pyximport; pyximport.install()
import permlib # This loads in the functions from permlib.pyx
import numpy as np; np.random.seed(7)
M = ortho_group.rvs(23) #Creates a random orthogonal matrix
%timeit permlib.npperm(M) # The numpy version
1 loop, best of 3: 44.5 s per loop
%timeit permlib.permfunc(M) # The cython version
1 loop, best of 3: 273 ms per loop
%timeit permlib.permfunc_modified(M) #romeric's improvement
10 loops, best of 3: 198 ms per loop
M = ortho_group.rvs(28)
%timeit permlib.permfunc(M) # The cython version run on a 28x28 matrix
1 loop, best of 3: 15.8 s per loop
%timeit permlib.permfunc_modified(M) # romeric's improvement run on a 28x28 matrix
1 loop, best of 3: 12.4 s per loop
Can the cython code be sped up at all?
I am using gcc and the CPU is the AMD FX 8350.
Upvotes: 8
Views: 2568
Answers (4)
Reputation: 2139
Disclaimer: I'm the core dev of the tool mentioned below.
As an alternative to Cython, you can give Pythran a try. A single annotation to the original NumPy code:
#pythran export npperm(float[:, :])
import numpy as np
def npperm(M):
n = M.shape[0]
d = np.ones(n)
j = 0
s = 1
f = np.arange(n)
v = M.sum(axis=0)
p = np.prod(v)
while j < n-1:
v -= 2*d[j]*M[j]
d[j] = -d[j]
s = -s
prod = np.prod(v)
p += s*prod
f[0] = 0
f[j] = f[j+1]
f[j+1] = j+1
j = f[0]
return p/2**(n-1)
compiled with:
> pythran perm.py
yields a speedup similar to the one with Cython:
> # numpy version
> python -mtimeit -r3 -n1 -s 'from scipy.stats import ortho_group; from perm import npperm; import numpy as np; np.random.seed(7); M = ortho_group.rvs(23)' 'npperm(M)'
1 loops, best of 3: 21.7 sec per loop
> # pythran version
> pythran perm.py
> python -mtimeit -r3 -n1 -s 'from scipy.stats import ortho_group; from perm import npperm; import numpy as np; np.random.seed(7); M = ortho_group.rvs(23)' 'npperm(M)'
1 loops, best of 3: 171 msec per loop
with no need to reimplement sum_axis (Pythran takes care of that).
More interesting, Pythran is capable of recognizing several vectorizable (in the sense of generating SSE/AVX intrinsics) patterns, through an option flag:
> pythran perm.py -DUSE_BOOST_SIMD -march=native
> python -mtimeit -r3 -n10 -s 'from scipy.stats import ortho_group; from perm import npperm; import numpy as np; np.random.seed(7); M = ortho_group.rvs(23)' 'npperm(M)'
10 loops, best of 3: 93.2 msec per loop
which makes a final x232 speedup with respect to the NumPy version, a speedup comparable to the unrolled Cython version, without much manual tuning.
Upvotes: 3
Reputation: 618
This answer is based on the code of @romeric posted before. I corrected the code and simplified it, and added the cdivisioncompiler directive.
@cython.boundscheck(False)
@cython.wraparound(False)
@cython.cdivision(True)
def permfunc_modified_2(np.ndarray [double, ndim =2, mode='c'] M):
cdef:
int n = M.shape[0], s=1, i, j
int *f = <int*>malloc(n*sizeof(int))
double *d = <double*>malloc(n*sizeof(double))
double *v = <double*>malloc(n*sizeof(double))
double p = 1, prod
for i in range(n):
v[i] = 0.
for j in range(n):
v[i] += M[j,i]
p *= v[i]
f[i] = i
d[i] = 1
j = 0
while (j < n-1):
prod = 1.
for i in range(n):
v[i] -= 2.*d[j]*M[j, i]
prod *= v[i]
d[j] = -d[j]
s = -s
p += s*prod
f[0] = 0
f[j] = f[j+1]
f[j+1] = j+1
j = f[0]
free(d)
free(f)
free(v)
return p/pow(2.,(n-1))
The original code of @romeric didn't initialize v, so you get different results sometimes. Additionally, I combined the two loops before the while and the two loops inside of the while respectively.
Finally, the comparison
In [1]: from scipy.stats import ortho_group
In [2]: import permlib
In [3]: import numpy as np; np.random.seed(7)
In [4]: M = ortho_group.rvs(5)
In [5]: np.equal(permlib.permfunc(M), permlib.permfunc_modified_2(M))
Out[5]: True
In [6]: %timeit permfunc(M)
10000 loops, best of 3: 20.5 µs per loop
In [7]: %timeit permlib.permfunc_modified_2(M)
1000000 loops, best of 3: 1.21 µs per loop
In [8]: M = ortho_group.rvs(15)
In [9]: np.equal(permlib.permfunc(M), permlib.permfunc_modified_2(M))
Out[9]: True
In [10]: %timeit permlib.permfunc(M)
1000 loops, best of 3: 1.03 ms per loop
In [11]: %timeit permlib.permfunc_modified_2(M)
1000 loops, best of 3: 432 µs per loop
In [12]: M = ortho_group.rvs(28)
In [13]: np.equal(permlib.permfunc(M), permlib.permfunc_modified_2(M))
Out[13]: True
In [14]: %timeit permlib.permfunc(M)
1 loop, best of 3: 14 s per loop
In [15]: %timeit permlib.permfunc_modified_2(M)
1 loop, best of 3: 5.73 s per loop
Upvotes: 1

Reputation: 2385
There isn't much you can do with your cython function, as it is already well optimised. However, you will still be able to get a moderate speed-up by completely avoiding the calls to numpy.
import numpy as np
cimport numpy as np
cimport cython
from libc.stdlib cimport malloc, free
from libc.math cimport pow
cdef inline double sum_axis(double *v, double *M, int n):
cdef:
int i, j
for i in range(n):
for j in range(n):
v[i] += M[j*n+i]
@cython.boundscheck(False)
@cython.wraparound(False)
def permfunc_modified(np.ndarray [double, ndim =2, mode='c'] M):
cdef:
int n = M.shape[0], j=0, s=1, i
int *f = <int*>malloc(n*sizeof(int))
double *d = <double*>malloc(n*sizeof(double))
double *v = <double*>malloc(n*sizeof(double))
double p = 1, prod
sum_axis(v,&M[0,0],n)
for i in range(n):
p *= v[i]
f[i] = i
d[i] = 1
while (j < n-1):
for i in range(n):
v[i] -= 2.*d[j]*M[j, i]
d[j] = -d[j]
s = -s
prod = 1
for i in range(n):
prod *= v[i]
p += s*prod
f[0] = 0
f[j] = f[j+1]
f[j+1] = j+1
j = f[0]
free(d)
free(f)
free(v)
return p/pow(2.,(n-1))
Here are essential checks and timings:
In [1]: n = 12
In [2]: M = np.random.rand(n,n)
In [3]: np.allclose(permfunc_modified(M),permfunc(M))
True
In [4]: n = 28
In [5]: M = np.random.rand(n,n)
In [6]: %timeit permfunc(M) # your version
1 loop, best of 3: 28.9 s per loop
In [7]: %timeit permfunc_modified(M) # modified version posted above
1 loop, best of 3: 21.4 s per loop
EDIT
Lets perform some basic SSE vectorisation by unrolling the inner prod loop, that is change the loop in the above code to the following
# define t1, t2 and t3 earlier as doubles
t1,t2,t3=1.,1.,1.
for i in range(0,n-1,2):
t1 *= v[i]
t2 *= v[i+1]
# define k earlier as int
for k in range(i+2,n):
t3 *= v[k]
p += s*(t1*t2*t3)
and now the timing
In [8]: %timeit permfunc_modified_vec(M) # vectorised
1 loop, best of 3: 14.0 s per loop
So almost 2X speed-up over the original optimised cython code, not bad.
Upvotes: 3
Reputation: 39039
Well, one obvious optimization is to set d[i] to -2 and +2 and avoid the multiplication by 2. I suspect this won't make any difference, but still.
Another is to make sure the C++ compiler that compiles the resulting code has all the optimizations turned on (especially vectorization).
The loop that calculates the new v[i]s can be parallelized with Cython's support of OpenMP. At 30 iterations this also might not make a difference.
Upvotes: 0
Related Questions
- Can I further optimize this Cython code snippet (contains NumPy and a string)?
- Is there a way of improving speed of cython code
- Further optimization of simple cython code
- Cython optimization of the code
- Making my cython code more efficient
- Speed-up cython code
- Speed up function using cython
- Speeding up my numpy code
- Speeding up Python Numpy code
- Speeding up python code with cython