Reputation: 31073
Bar plot with log scales
I've run into an interesting problem with scaling using ggplot. I have a dataset that I can graph just fine using the default linear scale but when I use scale_y_log10() the numbers go way off. Here is some example code and two pictures. Note that the max value in the linear scale is ~700 while the log scaling results in a value of 10^8. I show you that the entire dataset is only ~8000 entries long so something is not right.
I imagine the problem has something to do with the structure of my dataset and the binning as I cannot replicate this error on a common dataset like 'diamonds.' However I am not sure the best way to troubleshoot.
thanks, zach cp
Edit: bdamarest can reproduce the scale problem on the diamond dataset like this:
example_1 = ggplot(diamonds, aes(x=clarity, fill=cut)) +
geom_bar() + scale_y_log10(); print(example_1)
#data.melt is the name of my dataset
> ggplot(data.melt, aes(name, fill= Library)) + geom_bar()
> ggplot(data.melt, aes(name, fill= Library)) + geom_bar() + scale_y_log10()
> length(data.melt$name)
[1] 8003


here is some example data... and I think I see the problem. The original melted dataset may have been ~10^8 rows long. Maybe the row numbers are being used for the stats?
> head(data.melt)
Library name group
221938 AB Arthrofactin glycopeptide
235087 AB Putisolvin cyclic peptide
235090 AB Putisolvin cyclic peptide
222125 AB Arthrofactin glycopeptide
311468 AB Triostin cyclic depsipeptide
92249 AB CDA lipopeptide
test2 <- data.frame(
Library = rep("AB", 6L),
name = c(
"Arthrofactin", "Putisolvin", "Putisolvin", "Arthrofactin",
"Triostin", "CDA"
),
group = c(
"glycopeptide", "cyclic peptide", "cyclic peptide", "glycopeptide",
"cyclic depsipeptide", "lipopeptide"
),
row.names = c(221938L, 235087L, 235090L, 222125L, 311468L, 92249L)
)
UPDATE:
Row numbers are not the issue. Here is the same data graphed using the same aes x-axis and fill color and the scaling is entirely correct:
> ggplot(data.melt, aes(name, fill= name)) + geom_bar()
> ggplot(data.melt, aes(name, fill= name)) + geom_bar() + scale_y_log10()
> length(data.melt$name)
[1] 8003


Upvotes: 43
Views: 32706
Answers (2)
Reputation: 2079
The best option is to get rid of the bar stacking (as @Brian commented log(sum(x)) != sum(log(x))) by using facet_wrap. You could also add a panel to represent the Total if needed.
For instance, for the diamonds dataset (following @Brian Diggs's answer), we could either plot
diamonds %>%
bind_rows( # Adds a
diamonds %>% # panel to
mutate(cut = "Total") # represent
) %>% # the
mutate(cut = cut %>% fct_relevel("Total", after = Inf)) %>% # Total
ggplot(aes(x = clarity, fill = clarity)) +
geom_bar() +
facet_wrap(~cut) +
scale_y_log10()
or,
diamonds %>%
bind_rows( # Adds a
diamonds %>% # panel to
mutate(clarity = "Total") # represent
) %>% # the
mutate(clarity = clarity %>% fct_relevel("Total", after = Inf)) %>% # Total
ggplot(aes(x = cut, fill = cut)) +
geom_bar() +
facet_wrap(~clarity) +
scale_y_log10()
Upvotes: 2
Reputation: 58875
geom_bar and scale_y_log10 (or any logarithmic scale) do not work well together and do not give expected results.
The first fundamental problem is that bars go to 0, and on a logarithmic scale, 0 is transformed to negative infinity (which is hard to plot). The crib around this usually to start at 1 rather than 0 (since $\log(1)=0$), not plot anything if there were 0 counts, and not worry about the distortion because if a log scale is needed you probably don't care about being off by 1 (not necessarily true, but...)
I'm using the diamonds example that @dbemarest showed.
To do this in general is to transform the coordinate, not the scale (more on the difference later).
ggplot(diamonds, aes(x=clarity, fill=cut)) +
geom_bar() +
coord_trans(ytrans="log10")
But this gives an error
Error in if (length(from) == 1 || abs(from[1] - from[2]) < 1e-06) return(mean(to)) :
missing value where TRUE/FALSE needed
which arises from the negative infinity problem.
When you use a scale transformation, the transformation is applied to the data, then stats and arrangements are made, then the scales are labeled in the inverse transformation (roughly). You can see what is happening by breaking out the calculations yourself.
DF <- ddply(diamonds, .(clarity, cut), summarise, n=length(clarity))
DF$log10n <- log10(DF$n)
which gives
> head(DF)
clarity cut n log10n
1 I1 Fair 210 2.322219
2 I1 Good 96 1.982271
3 I1 Very Good 84 1.924279
4 I1 Premium 205 2.311754
5 I1 Ideal 146 2.164353
6 SI2 Fair 466 2.668386
If we plot this in the normal way, we get the expected bar plot:
ggplot(DF, aes(x=clarity, y=n, fill=cut)) +
geom_bar(stat="identity")
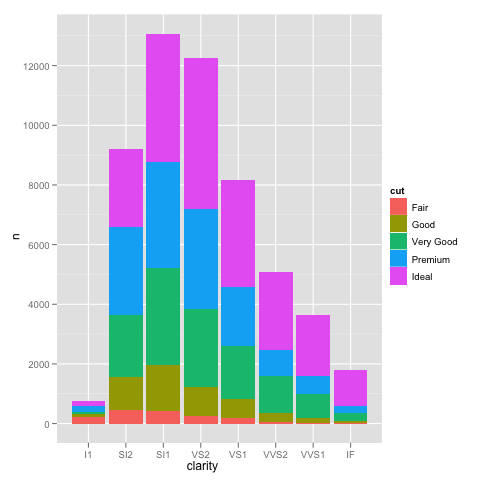
and scaling the y axis gives the same problem as using the not pre-summarized data.
ggplot(DF, aes(x=clarity, y=n, fill=cut)) +
geom_bar(stat="identity") +
scale_y_log10()

We can see how the problem happens by plotting the log10() values of the counts.
ggplot(DF, aes(x=clarity, y=log10n, fill=cut)) +
geom_bar(stat="identity")

This looks just like the one with the scale_y_log10, but the labels are 0, 5, 10, ... instead of 10^0, 10^5, 10^10, ...
So using scale_y_log10 makes the counts, converts them to logs, stacks those logs, and then displays the scale in the anti-log form. Stacking logs, however, is not a linear transformation, so what you have asked it to do does not make any sense.
The bottom line is that stacked bar charts on a log scale don't make much sense because they can't start at 0 (where the bottom of a bar should be), and comparing parts of the bar is not reasonable because their size depends on where they are in the stack. Considered instead something like:
ggplot(diamonds, aes(x=clarity, y=..count.., colour=cut)) +
geom_point(stat="bin") +
scale_y_log10()

Or if you really want a total for the groups that stacking the bars usually would give you, you can do something like:
ggplot(diamonds, aes(x=clarity, y=..count..)) +
geom_point(aes(colour=cut), stat="bin") +
geom_point(stat="bin", colour="black") +
scale_y_log10()

Upvotes: 56
Related Questions
- Bar notation in base-10 log scale in ggplot2
- ggplot log scale y axis messes up stat_summary bar plot
- Showing value of 1 in ggplot2 bar graph with log scale
- Create bar plot with logarithmic scale in R
- log-log scale y axis ggplot
- Trouble with log scale on ggplot grouped bar plot
- log scale error in r during barplot
- ggplot barplot : How to display small positive numbers with log scaled y-axis
- R ggplot log log scale
- ggplot in R: barchart with log scale label misplacement

